
Hi think you will never see a conventional list perform as fast as an array. Is there a better / faster way of using read-only Python lists (or even a list of tuples) with Numba jit than I have done above? Will eventually run just as fast with Python lists as Numpy arrays, so I can use a Python list or even a Python list of tuples directly with my Numba jit functions? Or will it most likely always be much faster to pass Numpy arrays to Numba jit functions? It seems that in the current version of Numba (0.54.1), the fastest method by far, is to pass Numpy arrays instead of Python lists to Numba jit functions. # This is very fast when using separate Numpy arrays as the arguments. %timeit sum_matrix_sparse2(x_col_list=x_col_np, x_row_list=x_row_np, x_val_list=x_val_np) # This runs without a warning but is much slower than 'sum_matrix_dense()'. %timeit sum_matrix_sparse2(x_col_list=(x_col_list), x_row_list=(x_row_list), x_val_list=(x_val_list)) # 5.69 ms ± 2.14 ms per loop (mean ± std. # 'x_col_list' of function 'sum_matrix_sparse2'. %timeit sum_matrix_sparse2(x_col_list=x_col_list, x_row_list=x_row_list, x_val_list=x_val_list) of 7 runs, 1000 loops sum_matrix_sparse2(x_col_list, x_row_list, x_val_list): # This runs without warnings but is much slower than 'sum_matrix_dense()'. %timeit sum_matrix_sparse1(x_list_tup=(x_list_tup)) # 24.7 ms ± 1.58 ms per loop (mean ± std.

# 'x_list_tup' of function 'sum_matrix_sparse1'. %timeit sum_matrix_sparse1(x_list_tup=x_list_tup) # but in my real use-case I need those as well. # This is a bit contrived because the indices i and j are ignored, of 7 runs, 100000 loops sum_matrix_sparse1(x_list_tup): X_list_tup = list(zip(x_col_list, x_row_list, sum_matrix_dense(x): # Convert to a list of tuples, which is the format I prefer in my use-case. # Numpy arrays for the column and row indices, and the matrix values. # Convert the sparse matrix to a dense matrix. X_sparse = (n, n, density=0.1, format='coo') # Generate a sparse matrix with random elements.
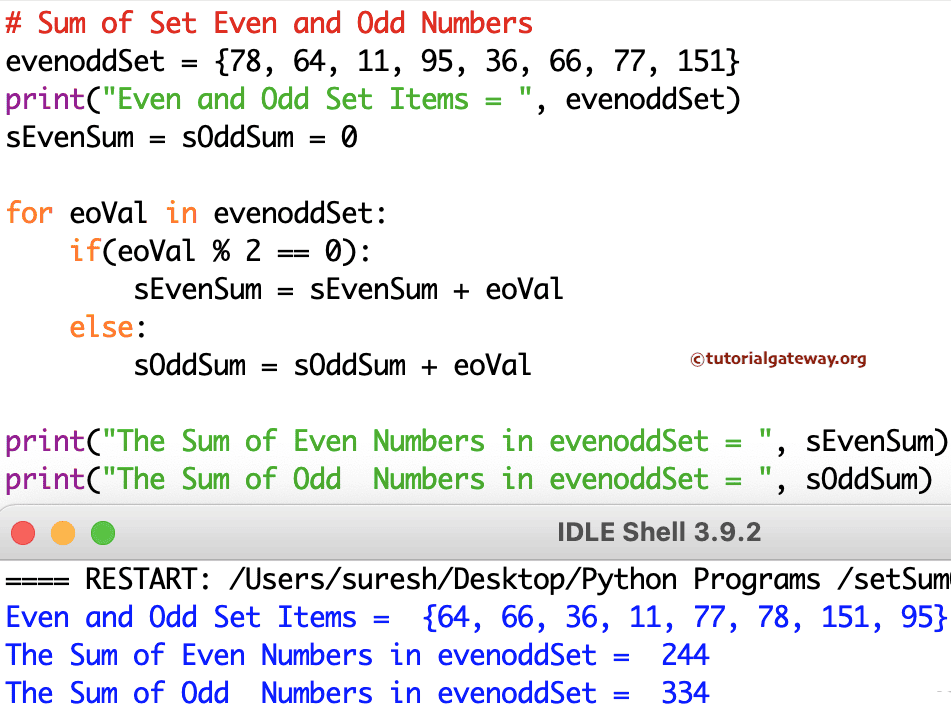
This does not seem to be supported very well in Numba as it runs very slowly.īelow is a somewhat contrived example which just sums all the elements of a sparse matrix and could therefore ignore the coordinates for the sparse matrix, but my actual problem needs to use the coordinates of the sparse matrix as well. In my actual use-case I have made a function that operates on a sparse matrix, whose elements are defined in a list of tuples like this: where the iX and jX are integer-values for the coordinates into the sparse matrix and vX are the float values for the matrix-elements at those coordinates. The slowest is to use the new and experimental. The fastest method by far is passing a Numpy array. # scheduled for deprecation: type 'reflected list' found for argument # NumbaPendingDeprecationWarning: Encountered the use of a type that is Let me first show the problem with a simple example, where we just sum all the elements of a list: # Number of elements in the list. My question relates to the best practices of passing read-only Python lists to Numba jit functions. In : C=sparse.Thanks for making Numba, I am a big fan! I’ve got the Numba fan-poster, t-shirt, cap, underwear and everything, even my bed-sheets say Numba! (OK, that’s not really true, but it could be!) And that list can have more than 2 arrays In : A=sparse.csr_matrix() All concatenate functions take a simple list or tuple of the arrays. np.hstack()Īnd don't use the double brackets. So you could just as well turned them into dense arrays (with.
One has 878048 nonzero elements - that means just one 0 element.
sparse matrices are not subclasses of numpy ndarray.īut, your 3 three matrices do not look sparse. As general rule you need to use sparse functions and methods, not the numpy ones with similar name. Therefore, How should I hstack a number of different sparse matrices?. Note that I am stacking such matrices since I would like to use them with a scikit-learn classification algorithm. However, the dimensions do not match: In : So I tried to hstack them: import numpy as np With 788618 stored elements in Compressed Sparse Row format>įrom the documentation, I read that it is possible to hstack, vstack, and concatenate them such type of matrices. With 744315 stored elements in Compressed Sparse Row format> With 878048 stored elements in Compressed Sparse Row format>


 0 kommentar(er)
0 kommentar(er)
